A new mathematical model of tumorigenesis—the formation of cancer—developed by scientists at the Johns Hopkins Kimmel Cancer Center, Whiting School of Engineering, and Bloomberg School of Public Health, suggests that the accumulation of random genetic mutations that launch a cell into malignancy later in life may get their start as early as the teen years. The model also offers an explanation for some longstanding mysteries about the timing and origin of cancer, such as why some cancer-causing mutations arise earlier or later in different varieties of this disease.
The findings were published December 23, 2019, in the Proceedings of the National Academy of Sciences.
“Current mathematical models of tumorigenesis are notoriously inaccurate,” says study leader Cristian Tomasetti, Ph.D., associate professor of oncology at the Johns Hopkins Kimmel Cancer Center. “Eventually, we may be able to help patients estimate their risk far more precisely, giving them an opportunity to act long before they’re in danger.”
He explains that the numerous mathematical models of tumor evolution developed to date often follow the rise of cancers beginning when the first cancer-causing mutation occurs, lending little insight into when that might take place over the course of a lifetime. And those models that do include the time before that first mutation, he says, often don’t produce predictions that match epidemiological data about cancer incidence well. In the rare instances when they provide reasonable approximations, it is by fitting the mathematical model to a single cancer type, not generalizing well to other cancers. The risk is to stray from what’s known about cancer biology to match these curves.
Tomasetti says that he and his colleague’s goal was to produce a new model that stayed as true to the current understanding of cancer biology as possible and see if it could closely match epidemiological data across several different cancer types. In fact, “it is relatively easy for a mathematical model to fit a curve, but not several different curves at once. With this new model we have been able to replicate the incidence of several cancer types at once, something never achieved before”. To do that, they designed their model to follow millions of cells and their divisions, a known source of cancer-causing mutations over time.
They included other key ingredients in the model, says Donald Geman, Ph.D., professor of applied mathematics and statistics at Johns Hopkins Whiting School of Engineering, including current understanding about the rate of random replicative genetic mutations that arise as cells make mistakes during copying, the number of stem cells, the only cells that are able to replicate themselves in an organ, and therefore with the potential to carry forward these mutations, and the rate of their division. They also included the limitations in resources that those cells must face and, to a degree never obtained before, the different effects that different types of cancer-causing mutations have on a cell.
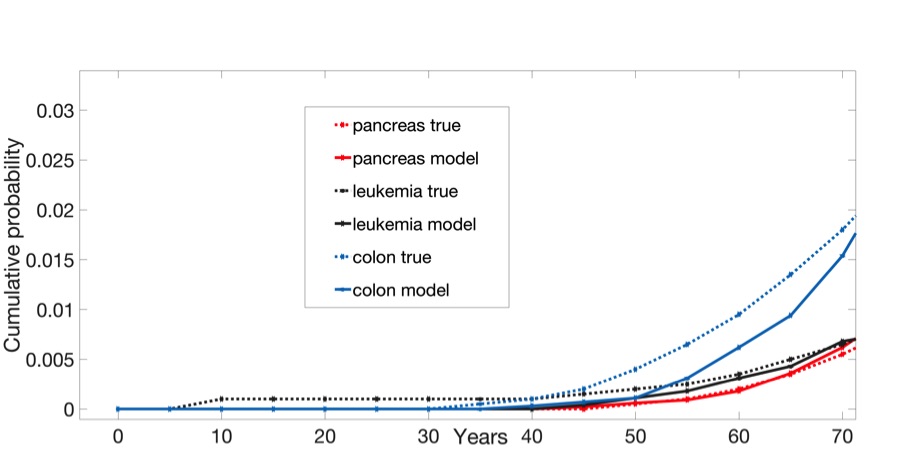
Cancer is considered a disease of aging with most diagnoses occurring after age 65. For this reason, the model covered an individual’s entire lifetime until cancer diagnosis: as organs were growing and reaching maturity in utero and over childhood, during homeostasis in adulthood when organs were full-grown and when enough cancer-driving mutations accumulate to classify cells as malignant, and until the resulting tumor grew large enough to be detectable. Using this model, they ran simulations for three relatively common and deadly cancers—colorectal, pancreatic, and leukemia—for 10,000 individuals apiece—and compared them with data obtained from the Surveillance, Epidemiology, and End Results (SEER) program of the National Cancer Institute.
For each of these cancers, the predicted incidence from the model provided a good match with true epidemiological data. For example, while the true mean of cancer detection from among individuals with an inherited form of colorectal cancer known as familial adenomatosis polyposis (FAP) is 39 years old, the estimated mean of the model was 41.33 years old.
The model also provides an explanation for why some mutations seem to be essential for spurring some cancers but less important for others. For example, a mutation in a gene known as KRAS is often one of the first mutations that kick off pancreatic cancer, but in colorectal cancer, this mutation typically occurs after other cancer-driving mutations get a foothold.
The answer, Tomasetti says, lies in the role of this gene and how quickly the cells in these organs divide. Because KRAS mutations can significantly speed up cell division, it has a stronger effect in the pancreas, where cells divide very slowly, about every eight months, than in the colon and rectum, where cells divide about every four days.
Intriguingly, the researchers say this model predicts that for the three cancers studied, the time point of the first mutation that will eventually lead to a cancer diagnosis is quite early in life (at 14.4 years of age for colorectal cancer, 17.4 years of age for leukemia, and 14.6 years of age for pancreatic cancer) with the full development of malignancy taking another 50 years or so on average. These findings are a significant deviation from estimates in previous research that put the time spans from first mutation to malignancy taking place in far shorter time periods—for example, in just seven years for leukemia.
The first author Kamel Lahouel, Ph.D., a Johns Hopkins Kimmel Cancer Center bioinformatics and biostatistics fellow, says that if further studies affirm that their model matches what's happening biologically, it could have enormous implications for helping people prevent and screen for cancers. For example, significant efforts are focused on sampling cells from distant organs through blood and other bodily fluids in tests known as liquid biopsies. If researchers were able to detect the rise of cancer-causing mutations early on, they might eventually be able to implement screening and other interventions that could prevent deadly late-stage malignancies. It could also help researchers fine-tune the timing of screenings, Lahouel says. Because such a long time span exists from when mutations begin to when cancer occurs, he explains, screening at different time points gives a long window to catch potential cancers before they are caught too late.
Remarkably, study researcher Laurent Younes, Ph.D., professor of applied mathematics and statistics at the Johns Hopkins Whiting School of Engineering, adds, the model produced data that closely matched incident data for all three cancers studied. The fact that this happened even without incorporating environmental factors that increase the mutation rate, such as smoking, suggests that background mutations may be responsible for a relevant fraction of cancer cases. He notes that he and his colleagues plan to continue to improve this model by incorporating other factors to make predictions even more accurate for individual circumstances.
Other Johns Hopkins researchers who participated in this study include Ludmila Danilova, Francis M. Giardiello, Ralph H. Hruban, John Groopman, Kenneth Kinzler and Bert Vogelstein.
This study was supported by The John Templeton Foundation, the Maryland Cigarette Restitution Fund, the Lustgarten Foundation, and the National Cancer Institute grant P30CA006973.